Integrative and perturbation based analysis of the transcriptional dynamics of TGFβ/BMP system components in transition from embryonic stem cells to neural progenitors.
Abstract
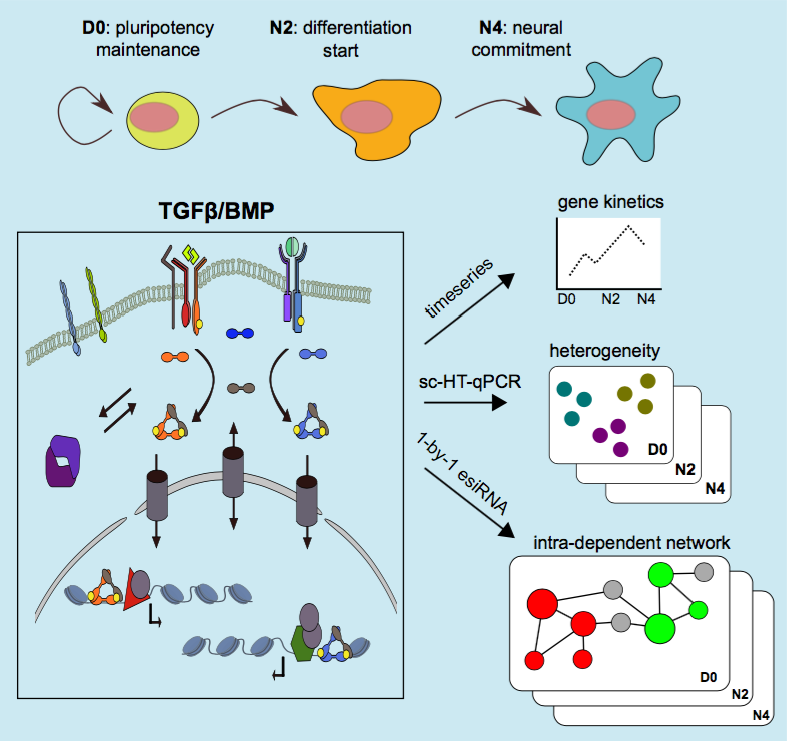
Cooperative actions of extrinsic signals and cell-intrinsic transcription factors alter gene regulatory networks enabling cells to respond appropriately to environmental cues. Signaling by Transforming Growth Factor type β (TGFβ) family ligands (eg, Bone Morphogenetic Proteins [BMPs] and Activins/Nodal) exerts cell-type specific and context-dependent transcriptional changes, thereby steering cellular transitions throughout embryogenesis. Little is known about coordinated regulation and tran- scriptional interplay of the TGFβ system. To understand intra-family transcriptional regulation as part of this system’s actions during development, we selected 95 of its components and investigated their mRNA-expression dynamics, gene-gene interac- tions and single-cell expression heterogeneity in mouse embryonic stem cells (mESCs) transiting to neural progenitors. Interrogation at 24 hour intervals identified 4 types of temporal gene transcription profile that capture all stages, that is, pluripotency, epi- blast formation and neural commitment. Then, between each stage we performed esiRNA-based perturbation of each individual component and documented the effect on steady-state mRNA levels of the remaining 94 components. This exposed an intri- cate system of multi-level regulation whereby the majority of gene-gene interactions display a marked cell-stage specific behavior. Furthermore, single-cell RNA-profiling at individual stages demonstrated the presence of detailed co-expression modules and subpopulations showing stable co-expression modules such as that of the core pluripotency genes at all stages. Our combinatorial experimental approach demon- strates how intrinsically complex transcriptional regulation within a given pathway is during cell fate/state transitions.
Contribution: Design, generation, processing, analyzing and interpretation of all data. Discussing, writing and editing of paper.